Jim: Difference between revisions
imported>Psych204B No edit summary |
imported>Psych204B No edit summary |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
= Background = | = Background = | ||
This quarter, we've talked a great deal about the importance of making deliberate choices at ever step of imaging analysis, and not trusting a 'black-box' pipeline. In this spirit, I decided to take a closer look at a typical preprocessing stream used in my lab. In doing so, I noticed variation in the preprocessing steps taken in my lab at even the earliest steps of preprocessing. Specifically, noticed some variation in the implementation of volume registration, and how decisions about the inputs to this volume registration can affect the motion parameters by this process. Accordingly, for my project, I decided to take a look at this variation. To do so I performed volume registration on previously collected imaging data from two different subjects, varying the chosen reference volume and the amount of data included in the imaging process. | This quarter, we've talked a great deal about the importance of making deliberate choices at ever step of imaging analysis, and not trusting a 'black-box' pipeline. In this spirit, I decided to take a closer look at a typical preprocessing stream used in my lab. In doing so, I noticed variation in the preprocessing steps taken in my lab at even the earliest steps of preprocessing. Specifically, noticed some variation in the implementation of volume registration, and how decisions about the inputs to this volume registration can affect the motion parameters by this process. Accordingly, for my project, I decided to take a look at this variation. To do so I performed volume registration on previously collected imaging data from two different subjects, varying the chosen reference volume and the amount of data included in the imaging process. | ||
<br> | |||
== Volume Registration == | == Volume Registration == | ||
An important early step of fMRI image preprocessing is volume registration. During scanning, there is inevitably some variance in the position of a participant’s brain. Participants (despite even the best intentions) move during a scan, and the brain will also move in the skull thanks to pulsation. Volume registration is one way of (partially) accounting for this. During volume registration, each acquired volume (full set of slices acquired in a single TR) is aligned to a template volume. During this process movement is estimated at each TR by measuring the displacement of an image from the reference image in 6 different dimensions, displacement in the x, y and z planes, and translation in terms of roll, pitch and yaw. These estimates are commonly used later in data analysis as nuisance parameters in the GLM (generalized linear model). | An important early step of fMRI image preprocessing is volume registration. During scanning, there is inevitably some variance in the position of a participant’s brain. Participants (despite even the best intentions) move during a scan, and the brain will also move in the skull thanks to pulsation. Volume registration is one way of (partially) accounting for this. During volume registration, each acquired volume (full set of slices acquired in a single TR) is aligned to a template volume. During this process movement is estimated at each TR by measuring the displacement of an image from the reference image in 6 different dimensions, displacement in the x, y and z planes, and translation in terms of roll, pitch and yaw. These estimates are commonly used later in data analysis as nuisance parameters in the GLM (generalized linear model). | ||
<br> | |||
= Methods = | = Methods = | ||
== Data Acquisition == | == Data Acquisition == | ||
Two female participants aged 40 and 50 were scanned as a part of an ongoing project studing intergenerational risk factors for depression. Scans were conducted on a 3T scanner at the Lucas Center. Scans were 314 Volumes long, with a TR of 2000 MS and acquired with a flip angle=70 degrees, TE = 40 msec, FOV 204 and a T2* weighted sprial in/out pulse sequence. Volumes were composed of 24 axial slices, 3.0 mm per slice, 3.75 x 3.75 mm in plane resolution. | |||
Two female participants aged 40 and 50 were scanned as a part of an ongoing project studing intergenerational risk factors for depression. Scans were conducted on a 3T scanner at the Lucas Center. Scans were 314 Volumes long, with a TR of 2000 MS and acquired with a flip angle=70 degrees, TE = 40 msec, FOV 204 and a T2* weighted sprial in/out pulse sequence. Volumes were composed of 24 axial slices, 3.0 mm per slice, 3.75 x 3.75 | |||
mm in plane resolution. | |||
== Data Analysis == | == Data Analysis == | ||
Data analysis was conducted using AFNI (Analysis of Functional NeuroImaging), a data analysis program developed by Bob Cox. To test factors affecting the motion parameters, volume registration was conducted varying two inputs of interest. The first was the input data; during fMRI acquisition, the MR scanner typically takes several TRs to reach equilibrium. While the data acquired during the first few TRs is typically discarded prior to image analysis; some preprocessing scripts trim this initial TRs later in preprocessing. Accordingly, I conducted volume registration on both a full data set that included these initial TRs, and a trimmed data set that included only the ‘settled’ TRs. | Data analysis was conducted using AFNI (Analysis of Functional NeuroImaging), a data analysis program developed by Bob Cox. To test factors affecting the motion parameters, volume registration was conducted varying two inputs of interest. The first was the input data; during fMRI acquisition, the MR scanner typically takes several TRs to reach equilibrium. While the data acquired during the first few TRs is typically discarded prior to image analysis; some preprocessing scripts trim this initial TRs later in preprocessing. Accordingly, I conducted volume registration on both a full data set that included these initial TRs, and a trimmed data set that included only the ‘settled’ TRs. | ||
Additionally, I also varied the volume chosen as the template volume for registration. For each data set (‘trimmed’ and ‘settled’), I conducted volume registration using either the initial (0), 90th or 300th TR of each data set as the template volume. These data analyses were performed for each subject. The 90th and initial volumes were chosen because they were both options chosen in preprocessing scripts for our lab, the later TR was chosen as it seemed interesting. These analyses were conducted on both participants’ data. The outcome measures for these analyses were the maximum displacement a volume as provided by 3dVolReg in afni, as well as the variance, means, and correlation (across analysis choice) of the 6 motion parameters produced by volume registration. I hypothesized that Volume registration conducted on the full data set with the initial TR as the reference volume would produce the worst motion parameters (highest variance, greatest motion displacement), but wanted to investigate whether including the initial TRs would distort motion parameters even when a later (90,300) TR was chosen as the reference volume. | |||
Additionally, I also varied the volume chosen as the template volume for registration. For each data set (‘trimmed’ and ‘settled’), I conducted | <br> | ||
volume registration using either the initial (0), 90th or 300th TR of each data set as the template volume. These data analyses were performed for each subject. The 90th and initial volumes were chosen because they were both options chosen in preprocessing scripts for our lab, the later TR was chosen as it seemed interesting. These analyses were conducted on both participants’ data. The outcome measures for these analyses were the maximum displacement a volume as provided by 3dVolReg in afni, as well as the variance, means, and correlation (across analysis choice) of the 6 motion parameters produced by volume registration. I hypothesized that Volume registration conducted on the full data set with the initial TR as the reference volume would produce the worst motion parameters (highest variance, greatest motion displacement), but wanted to investigate whether including the initial TRs would distort motion parameters even when a later (90,300) TR was chosen as the reference volume. | |||
= Results = | = Results = | ||
Below are summary statistics from volume registration. Several things pop out from this data Analysis. The first is that there seems to be appreciable variance between participants. Subject 1 seems to have moved more – regardless of data set or reference volume, Subject one has greater variance, maximum displacement and mean value of motion parameters than Subject 2. Additionally, it seems clear that both chosen data set and template volume affect parameter estimates. Looking at the first column, there seems to be the greatest maximum displacement for both subjects for the full dataset fcompared to the trimmed ‘settled only’ data set. Additionally, for both subjects the 90th TR produced motion parameters with the smallest maximum displacement. | |||
[[File:Summary Statistics.png |thumb|400px|center| Summary Statistics]] | |||
<br> | <br> | ||
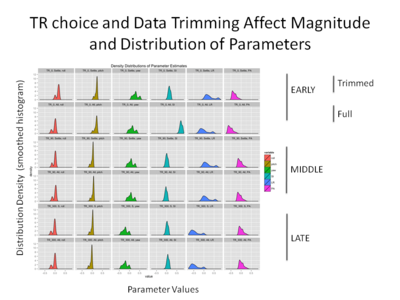
An examination of the distribution of motion parameters (see below) supports our initial intuition that both data set and reference volume affect motion parameters, though there does seem to be considerable overlap- the distributions of the various parameters seem to be of similar shapes across analyses, and seem to be roughly in the same place on the x-axis. Moreover, we can see that the motion parameters generated using the full data set with the initial TR as the reference volume seem to be the most deviant. | |||
[[File: | [[File: Distribution of mvmt parameters.png |thumb|400px|center| Smoothed Density Distribution of Motion parameters for Subject 1, split by data set used and reference TR]] | ||
<br> | <br> | ||
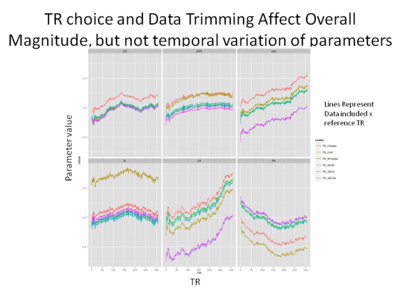
In the final figure below, motion parameters of Subject 1 are charted out as a function of TR (x-axis) and magnitude (y-axis). The parameters are split such that each panel represents an individual motion parameter (ie roll or pitch), and the various lines indicate the data set and reference TRs chosen. Here we see that there seems to be considerable variation in the magnitude of the parameters- for instance the lines tracking the ‘yaw’ parameter seem to be quite far apart for the various analyses. However, it seems clear that the differences only appear to be a main effect- across analyses the parameters appear to be quite tightly correlated, rising and falling quite closely. Indeed, after performing a computing a correlation matrix, I found that within parameters, the correlation across analyses was generally extremely high (~.99). This would indicate that although there may be some variation in the parameters, these differences may only be trivial and therefore not have a large influence on later analysis (say during GLM when they are included as nuisance regressors) | |||
[[File: TR split.png |thumb|400px|center| Magnitude of motion parameters plotted by TR, split by parameter type, data set included and reference TR chosen.]] | |||
[[File: | |||
<br> | <br> | ||
= Conclusions = | |||
Altogether, it seems that both the data set and the TR chosen as a reference point can influence the values of the motion parameters estimated during volume registration. However, it seems that these differences are largely ones of magnitude, and may not reflect temporal changes. It seems that the most deviant parameters are generated by volume registration which includes a data set with the initial TRs (during which the scanner is still settling down), and has the intial TR as the template volume. Additionally, it seems that including these initial TRs can influence the magnitude of motion parameters even when a later TR is chosen as the reference volume. Altogether, it seems that if one is seeking to minimize variation in motion parameters, the best practice is to trim one’s data set prior to registration, and to choose a reference volume that is relatively early in a run (but not one acquired during the first useable TR). | |||
Latest revision as of 11:15, 17 March 2012
Background
This quarter, we've talked a great deal about the importance of making deliberate choices at ever step of imaging analysis, and not trusting a 'black-box' pipeline. In this spirit, I decided to take a closer look at a typical preprocessing stream used in my lab. In doing so, I noticed variation in the preprocessing steps taken in my lab at even the earliest steps of preprocessing. Specifically, noticed some variation in the implementation of volume registration, and how decisions about the inputs to this volume registration can affect the motion parameters by this process. Accordingly, for my project, I decided to take a look at this variation. To do so I performed volume registration on previously collected imaging data from two different subjects, varying the chosen reference volume and the amount of data included in the imaging process.
Volume Registration
An important early step of fMRI image preprocessing is volume registration. During scanning, there is inevitably some variance in the position of a participant’s brain. Participants (despite even the best intentions) move during a scan, and the brain will also move in the skull thanks to pulsation. Volume registration is one way of (partially) accounting for this. During volume registration, each acquired volume (full set of slices acquired in a single TR) is aligned to a template volume. During this process movement is estimated at each TR by measuring the displacement of an image from the reference image in 6 different dimensions, displacement in the x, y and z planes, and translation in terms of roll, pitch and yaw. These estimates are commonly used later in data analysis as nuisance parameters in the GLM (generalized linear model).
Methods
Data Acquisition
Two female participants aged 40 and 50 were scanned as a part of an ongoing project studing intergenerational risk factors for depression. Scans were conducted on a 3T scanner at the Lucas Center. Scans were 314 Volumes long, with a TR of 2000 MS and acquired with a flip angle=70 degrees, TE = 40 msec, FOV 204 and a T2* weighted sprial in/out pulse sequence. Volumes were composed of 24 axial slices, 3.0 mm per slice, 3.75 x 3.75 mm in plane resolution.
Data Analysis
Data analysis was conducted using AFNI (Analysis of Functional NeuroImaging), a data analysis program developed by Bob Cox. To test factors affecting the motion parameters, volume registration was conducted varying two inputs of interest. The first was the input data; during fMRI acquisition, the MR scanner typically takes several TRs to reach equilibrium. While the data acquired during the first few TRs is typically discarded prior to image analysis; some preprocessing scripts trim this initial TRs later in preprocessing. Accordingly, I conducted volume registration on both a full data set that included these initial TRs, and a trimmed data set that included only the ‘settled’ TRs.
Additionally, I also varied the volume chosen as the template volume for registration. For each data set (‘trimmed’ and ‘settled’), I conducted volume registration using either the initial (0), 90th or 300th TR of each data set as the template volume. These data analyses were performed for each subject. The 90th and initial volumes were chosen because they were both options chosen in preprocessing scripts for our lab, the later TR was chosen as it seemed interesting. These analyses were conducted on both participants’ data. The outcome measures for these analyses were the maximum displacement a volume as provided by 3dVolReg in afni, as well as the variance, means, and correlation (across analysis choice) of the 6 motion parameters produced by volume registration. I hypothesized that Volume registration conducted on the full data set with the initial TR as the reference volume would produce the worst motion parameters (highest variance, greatest motion displacement), but wanted to investigate whether including the initial TRs would distort motion parameters even when a later (90,300) TR was chosen as the reference volume.
Results
Below are summary statistics from volume registration. Several things pop out from this data Analysis. The first is that there seems to be appreciable variance between participants. Subject 1 seems to have moved more – regardless of data set or reference volume, Subject one has greater variance, maximum displacement and mean value of motion parameters than Subject 2. Additionally, it seems clear that both chosen data set and template volume affect parameter estimates. Looking at the first column, there seems to be the greatest maximum displacement for both subjects for the full dataset fcompared to the trimmed ‘settled only’ data set. Additionally, for both subjects the 90th TR produced motion parameters with the smallest maximum displacement.

An examination of the distribution of motion parameters (see below) supports our initial intuition that both data set and reference volume affect motion parameters, though there does seem to be considerable overlap- the distributions of the various parameters seem to be of similar shapes across analyses, and seem to be roughly in the same place on the x-axis. Moreover, we can see that the motion parameters generated using the full data set with the initial TR as the reference volume seem to be the most deviant.
In the final figure below, motion parameters of Subject 1 are charted out as a function of TR (x-axis) and magnitude (y-axis). The parameters are split such that each panel represents an individual motion parameter (ie roll or pitch), and the various lines indicate the data set and reference TRs chosen. Here we see that there seems to be considerable variation in the magnitude of the parameters- for instance the lines tracking the ‘yaw’ parameter seem to be quite far apart for the various analyses. However, it seems clear that the differences only appear to be a main effect- across analyses the parameters appear to be quite tightly correlated, rising and falling quite closely. Indeed, after performing a computing a correlation matrix, I found that within parameters, the correlation across analyses was generally extremely high (~.99). This would indicate that although there may be some variation in the parameters, these differences may only be trivial and therefore not have a large influence on later analysis (say during GLM when they are included as nuisance regressors)
Conclusions
Altogether, it seems that both the data set and the TR chosen as a reference point can influence the values of the motion parameters estimated during volume registration. However, it seems that these differences are largely ones of magnitude, and may not reflect temporal changes. It seems that the most deviant parameters are generated by volume registration which includes a data set with the initial TRs (during which the scanner is still settling down), and has the intial TR as the template volume. Additionally, it seems that including these initial TRs can influence the magnitude of motion parameters even when a later TR is chosen as the reference volume. Altogether, it seems that if one is seeking to minimize variation in motion parameters, the best practice is to trim one’s data set prior to registration, and to choose a reference volume that is relatively early in a run (but not one acquired during the first useable TR).