Stephanie: Difference between revisions
imported>Psych204B Created page with '<br> = Background = LKcj Jaskdkghfls kjhkjcvx,mvkjifdjvjkrngkjv To add an image, simply put text like this inside double brackets 'MyFile.jpg | My figure caption'. When you s…' |
imported>Psych204B No edit summary |
||
| Line 19: | Line 19: | ||
=== Subjects === | === Subjects === | ||
Subjects were 19 healthy volunteers. | Subjects were 19 healthy, right-handed volunteers. | ||
== Memory Task == | == Memory Task == | ||
| Line 33: | Line 33: | ||
== fMRI Analysis == | == fMRI Analysis == | ||
=== MR acquisition === | === MR acquisition === | ||
Imaging data were acquired on a 3.0 T Signa whole-body MRI system with a custom-built head coil (GE Medical Systems, Milwaukee, WI, USA). | |||
=== | In total, 2,400 functional volumes were acquired for each participant using a T2*-sensitive gradient echo spiral in/our pulse sequence (Glover and Law, 2001). Functional imaging parameters were optimized to provide whole brain coverage (TR=2000ms; TE= 30ms; flip angle = 75°; FOV = 22 cm; 3.44 x 3.44 x 4 mm resolution, 30 slices). | ||
The | |||
=== fMRI Analysis === | |||
The fMRI data was analyzed using [https://github.com/mwaskom/lyman Lyman], [http://nipy.sourceforge.net/nipype/ Nipype], [https://surfer.nmr.mgh.harvard.edu/ Freesurfer], [http://afni.nimh.nih.gov/afni/ AFNI], and [http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/ FSL] software tools. | |||
==== Pre-processing ==== | ==== Pre-processing ==== | ||
All data | All data went through a standard preprocessing pipeline using Freesurfer and FSL, including motion (RapidART) and slice time correction, realignment (middle volume of each run), skull stripping, temporal filtering (high-pass cutoff 128 Hz), and surface-based coregistration (bbregister). Data were spatially smoothed (6 fwhm, SUSAN -- only averages a given voxel with local voxels that have a similar intensity), scaled grand median of timeseries to 10000, & normalized for group analyses (nonlinear warp to FSL’s MNI152 space). Data were modeled using a double gamma function, and the first 5 frames of each run were discarded. | ||
= Results = | = Results = | ||
Revision as of 05:00, 29 May 2013
Background
LKcj Jaskdkghfls kjhkjcvx,mvkjifdjvjkrngkjv
To add an image, simply put text like this inside double brackets 'MyFile.jpg | My figure caption'. When you save this text and click on the link, the wiki will ask you for the figure.
lKSJ Dkdjfdsklj lkfj cvlkxcvlkjes;lfjewlkj
Heading1
KJS Cj fijlckxzcjioe.lkcmxzicj
Materials and Methods
Subjects
Subjects were 19 healthy, right-handed volunteers.
Memory Task
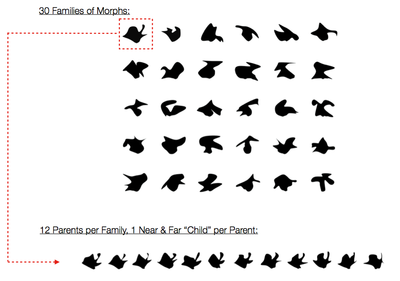
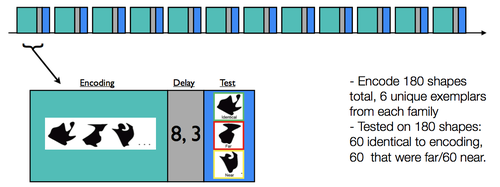
Behavioral Analysis
XXXXkdaldkjsalkdjas ;lkjsalkdjaslkdjaslk cjaslkfjaslkd jsalkdj alskdj alksdj alksdj aldkj aslkdj acmxkcjuifqw;l
fMRI Analysis
MR acquisition
Imaging data were acquired on a 3.0 T Signa whole-body MRI system with a custom-built head coil (GE Medical Systems, Milwaukee, WI, USA).
In total, 2,400 functional volumes were acquired for each participant using a T2*-sensitive gradient echo spiral in/our pulse sequence (Glover and Law, 2001). Functional imaging parameters were optimized to provide whole brain coverage (TR=2000ms; TE= 30ms; flip angle = 75°; FOV = 22 cm; 3.44 x 3.44 x 4 mm resolution, 30 slices).
fMRI Analysis
The fMRI data was analyzed using Lyman, Nipype, Freesurfer, AFNI, and FSL software tools.
Pre-processing
All data went through a standard preprocessing pipeline using Freesurfer and FSL, including motion (RapidART) and slice time correction, realignment (middle volume of each run), skull stripping, temporal filtering (high-pass cutoff 128 Hz), and surface-based coregistration (bbregister). Data were spatially smoothed (6 fwhm, SUSAN -- only averages a given voxel with local voxels that have a similar intensity), scaled grand median of timeseries to 10000, & normalized for group analyses (nonlinear warp to FSL’s MNI152 space). Data were modeled using a double gamma function, and the first 5 frames of each run were discarded.
Results
Behavioral Results
First, kljskdjf s;lkjsdflk kdjsf ksdfjklsdjf kjl
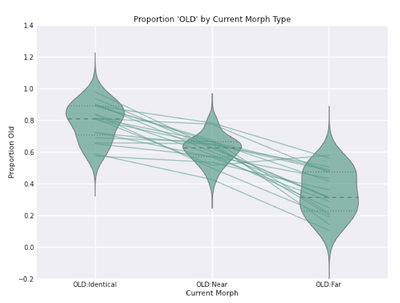
Then, skldjf klfjsdkfsjkcjxv;lcxkjvd dprimes...

K Jlkdsjfdslkfjsdlkfjds kjsdf lkasdjf;alsdkfj alsdkfj lskdjf
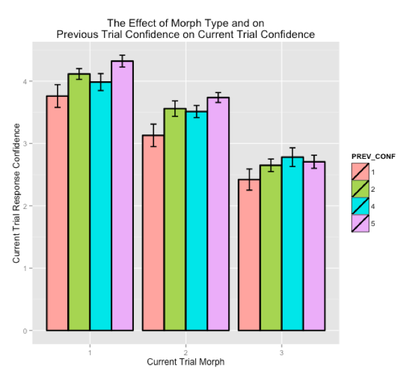
fMRI Results
KKLJkldjf skfjsdklfj kfj dsf;ls
Coding Regressors
Highpass Filter Cutoff
Conclusions
In summary, k jfklasjfdskaljd lksajdlksa jlksd
References
Software