Murthy: Difference between revisions
imported>Student221 |
imported>Student221 |
||
| Line 26: | Line 26: | ||
=== Generating Cone Mosaics === | === Generating Cone Mosaics === | ||
In order to execute the process depicted above, it was first necessary to generate the cone mosaics that would be used. For this project, it was desired to model two portions of the retina. For each model, a rectangular cone mosaic was generated using the “coneMosaic” method in ISETBio. | In order to execute the process depicted above, it was first necessary to generate the cone mosaics that would be used. For this project, it was desired to model two portions of the retina- the center of the fovea, and slightly to the periphery. For each model, a rectangular cone mosaic was generated using the “coneMosaic” method in ISETBio. | ||
First the “center” model was created. This cone mosaic is meant to represent the photoreceptors at the center of the fovea. Using the knowledge from Foundations of Vision that the S cones are absent at the center of the fovea [6], this model was created by specifying an even distribution of L and M cones, with no S cones and no blank spaces. The “coneMosaic” method in ISETBio defaults to generating cones with an aperture that is representative of the center of the fovea, so no special parameters needed to be set with respect to aperture. In this model, the aperture size was 1.4 micron. The “center” cone mosaic is depicted in Figure NUMBER below. | First the “center” model was created. This cone mosaic is meant to represent the photoreceptors at the center of the fovea. Using the knowledge from Foundations of Vision that the S cones are absent at the center of the fovea [6], this model was created by specifying an even distribution of L and M cones, with no S cones and no blank spaces. The “coneMosaic” method in ISETBio defaults to generating cones with an aperture that is representative of the center of the fovea, so no special parameters needed to be set with respect to aperture. In this model, the aperture size was 1.4 micron. The “center” cone mosaic is depicted in Figure NUMBER below. | ||
Revision as of 07:48, 13 December 2019
Introduction
The study of human eye movements and vision is an active research field. Over the past 35 years, researchers have focused on a few particular topics of interest, namely smooth pursuit, saccades, and the interactions of these movements with vision. The fundamental question of “how does the motion of the image on the retina affect vision?” has been a continuous area of focus.[1]
This knowledge of how eye movements impact vision has a wide variety of beneficial implications, from augmenting understanding of human behavior to assisting in the development of therapeutics. Eye movement studies have been used since the mid nineteenth century to understand human disease [2], and applications of eye movement research have the potential to alleviate vision disorders (such as the Stanford Artificial Retina Project, which can utilize such research to inform the prosthesis design).[3]
The study of eye movements and their impact on visual acuity can be challenging to conduct in vivo. Noise can be introduced from uncontrollable oculomotor muscle contractions, and it is challenging with human participants to perform repeatable eye movements and obtain an objective measure of their effect on vision. The ISETBio toolbox provides comprehensive functionality to enable the study of human visual acuity in a controlled simulation environment.[4]
The purpose of this project is to modify the eye movement patterns and cone properties of an ISETBio visual model, and observe trends related to the effects of these parameters on the visual acuity of the model. For this project, the Modulation Transfer Function (MTF) is used as an evaluation of visual acuity. The MTF is a curve depicting contrast reduction vs. spatial frequency, and it is a widely accepted research and industry standard method of comparing the performance of two visual systems.[5]
Background
Methods
MTF Computational Pipeline
ISO12233 provides a method for computing the MTF in a fast, standard manner. The MTF is, by definition, the magnitude of the Fourier Transform of the Linespread Function of the optical system. In turn, the Linespread Function is the derivative of the system’s response to an edge.[NUMBER] Using ISETBio and its ISO12233 function, the MTF can be generated by presenting a slanted edge to the visual model, then performing the aforementioned calculations to get from the edge response to the MTF curve.
In this regard, all MTFs for this project were generated via the computational pipeline depicted in Figure NUMBER. First, the “slanted edge” scene is generated. A macular pigment is applied to obtain the optical image of the scene. Second, a cone mosaic is created to produce the retina model. This process is described in more detail below. Third, the eye movement sequence is generated. This process is also described in more detail below. Fourth, the optical image is presented to the cone mosaic. The photon absorptions of each cone in the mosaic are computed for each time step (in this case, 5 milliseconds). Every time step, the presented image is shifted in accordance with the single eye movement for that time step. Finally, the mean absorptions for the cone mosaic are used to generate the MTF via the ISO12233 function. This process is repeated for the desired number of trials, producing one MTF per trial.
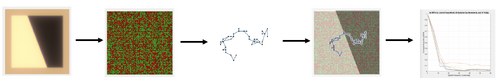
Generating Cone Mosaics
In order to execute the process depicted above, it was first necessary to generate the cone mosaics that would be used. For this project, it was desired to model two portions of the retina- the center of the fovea, and slightly to the periphery. For each model, a rectangular cone mosaic was generated using the “coneMosaic” method in ISETBio.
First the “center” model was created. This cone mosaic is meant to represent the photoreceptors at the center of the fovea. Using the knowledge from Foundations of Vision that the S cones are absent at the center of the fovea [6], this model was created by specifying an even distribution of L and M cones, with no S cones and no blank spaces. The “coneMosaic” method in ISETBio defaults to generating cones with an aperture that is representative of the center of the fovea, so no special parameters needed to be set with respect to aperture. In this model, the aperture size was 1.4 micron. The “center” cone mosaic is depicted in Figure NUMBER below.
Second, the “periphery” model was created. This cone mosaic is intended to represent the photoreceptors at a few degrees eccentricity relative to the center of the fovea. Initially, I considered making a mosaic that included blank spaces, which would represent rods. However, when attempting to run the ISO12233 routine using a model with blank cones, the resultant MTFs appeared to have erroneous shapes, indicating computational difficulty.
To address this issue while still attempting to generate a relatively accurate retinal model, the periphery model was chosen to represent 2 degrees of eccentricity. This decision was based on Figure 3.1 from Foundations of Vision (linked here) [6], which depicts the density of rods and cones as one moves across the retina. It is clear to see that while cone density falls off rapidly as one moves peripherally from the fovea, there is still a high concentration of cones at 2 degrees of eccentricity. To determine the aperture size for these cones, the ISETBio function “coneSizeReadData” was used. This function uses data from Curcio et. al. Figure 6, below, to estimate cone aperture size from cone density data.[NUMBER] Using this method, the cone aperture size for the periphery model was 3.89 micron. This mosaic is also depicted in Figure NUMBER below.