Mazzochette
Introduction:
In the Microsystems Lab, we are studying the sense of touch, specifically how our bodies undergo mechanotransduction, the process in which mechanical signals such as forces and displacements are converted into electrochemical signals that our nervous system can understand. The human body is incredibly complex, so we are first tackling the touch sensation problem in a popular neuroscience model organism, the Caenorhabditis elegans (C. elegans).
C. elegans is a small nematode, about 1 mm in length and about 50 um wide. It consists of about 1000 somatic cells and exactly 302 neurons. It was the first animal to have its genome fully mapped, as well as its complete neural network. Finally, it is a convenient sample in a biology lab because it grows to become a full adult animal in only four days. Because of all these traits, the animal lends itself easily to genetic mutation, allowing us to modulate different aspects of the worm’s composition for study.
 Image adapted from Goodman 2006
Image adapted from Goodman 2006
We know that the worm has exactly six neurons that respond to gentle touch sensation. The morphology of these neurons is consistent across the species, with three of the cell bodies located near the animal’s tail and the other three cell bodies located about 50% of the length of the body from the head. Each of the neurons has a long process that runs from the cell body towards the head (Goodman 2006). There exists strong evidence that points to specific ion channels in the processes of the neurons that open and close as strain is applied to the neuron. (O'Hagan 2004) As those channels open and close, ions flow in and out of the cell, creating a change in electric potential.
One method we are using to characterize the sense of touch is to quantify the behavioral response of the C. elegans to gentle touch. To do this, we probe the body of the worm with nano- and micro-scale forces and score whether or not the worm responds. We have built micro-scale cantlievers with an embedded strain gauge to apply calibrated forces and integrated them into a clamping system (Park 2011). We apply forces to a freely moving worm and observe the response through a video taken through a stereoscope. A behavioral response is scored positively when the worm’s trajectory changes immediately after a force application (Petzold 2013).
Currently, this system can apply forces with high precision, but the low spatial and temporal precision of targeting and analyzing behavioral response limits the data throughput and quality. Targeting of the worm is done semi-manually and the behavioral response is scored visually frame by frame. The figure below shows the manual intervention done on each frame so that the software can differentiate the worm from the cantilever. The user must actually drawn the white lines that are the boundary between the worm and the cantilever. The user must also specify the threshold used to find the skeleton indicated by the dashed line along the worm.
I am currently building a second generation of the system to enable automated tracking of the worm's skeleton in real time. With automated tracking we can precisely target a location along the body of the worm, such as 25% of the length of the skeleton from the head, as well as automate the behavioral response scoring. In this project I seek to design a more robust version of the hardware system as well as investigate methods for thresholding the images, the first step in the image processing. Acquiring images and thresholding are the first two steps in the process of tracking the worm. It is imperative that they be robust in order to accurately find the worm skeleton and target later in the processing pipeline.
Prior to starting this project, I already had a working prototype for acquiring a data set of video and images, as well as a basis code to skeletonize the worm. The hardware prototype consists primarily of a chemistry stand with a ring attached to it to hold the sample. The camera is placed below the sample. A light is held to the chemistry stand using a clamp. After capturing the images of the worm, I have written code based on the algorithm developed in (Leifer 2011) to segment the body of the worm, use the segments to skeletonize the worm and find specified target along the skeleton. The basis for my code development is a library of C++ functions called OpenCV, an open source computer vision platform that employs many novel algorithms in computer vision as well as a set of functions, classes and structures for computation with images.
Methods:
The success of high spatial tracking precision relies heavily on the front end image processing steps, namely taking a good image and thresholding smartly. In this project I hope to improve upon the original methods that I set out to use for these steps. The key metrics of a successful threshold are:
- Smooth outline of the worm's body
- The worm's body must be outlined by the longest contour of any feature deemed to be in the foreground by the thresholding method
- The quality of the images must be repeatable
- To enable real time processing, the time to process the image must be well below 66 ms, as governed by the 15 fps capture rate of the camera.
With a stable construction of the optics and smart thresholding, the I can create robust front end processsing, improving the accuracy and precision of locating a target on the body of the freely moving worm.
Part 1: Designing stable hardware
In the first part of this investigation, I hope to design a more robust hardware system so that images can be taken with repeatable quality and control. The first step in image processing, if controllable, is to take high quality images. I have found through taking several videos with my current system that stability in the construction and repeatable light placement is imperative for good analysis of images.
My imaging system has two key innovations. The first is an inverted configuration, where the light is collected from below the sample. The second is an obliquely oriented lighting source. This configuration leverages the relative optical properties of the C. elegans and the silicon, the material from which the cantilevers are made. The nematode will scatted the light as it enters the body of the worm, whereas the cantilever reflects light. With an oblique source, the light incident on the cantilever will reflect at such an oblique angle that it will not be captured by the optics below. The light incident on the worm however will be scattered in many directions, including downwards where the camera can capture it. The figure below shows a sketch of the system, as well as a set of sample images taken with a prototype of the system with two orientations of the light, above the sample and to the side of the sample.
A prototype of this system was previously built. Below is a sample video taken on the system, as well as a video of processed images from the first revision of the software. The faint outline of the cantilever can be seen in the upper left corner of the image. The noise generated by the cantilever is large enough to create an object in the foreground of the image with a larger contour than the worm. The processing mis-identifies the worm in almost every frame. An improved hardware system as well as better thresholding techniques will improve this processing. By looking at the strengths and weaknesses of different thresholding techniques, I intend to improve my design.
Part 2: Smart Thresholding
The first step in processing the images is to threshold to find the outline of the worm. In thresholding, all the pixels in the image are classified as either foreground (maxValue = 255) or background (minValue = 0) based on specified threshold. All thresholding methods follow the same basic structure as shown in the following equation. All techniques are structured around smartly picking .
As shown in the images above, some of the images include faint outline of the cantilever, which must be classified as background. In my original pass at the code, I arbitrarily guessed at thresholds until the images correctly divided into classes such that when contours were found, the largest contour was the worm. In this particular video, I had to crop out the cantilever to be able to find and process the worm.
In order to build a more robust processing algorithm, active thresholding techniques must be employed. To achieve this, I investigated three advanced methods for choosing T(x,y) such that body of the worm is designated as the picture foreground, while everything else in the image is specified as background. The first method is a popular, stochastically based algorithm called Otsu’s Method. The second and third methods are adaptive threshold techniques where a mean kernel and Gaussian kernel are used to determine a local threshold. All three of these methods are built into the OpenCV image processing library, simplifying implementation.
Otsu’s Method
The goal of Otsu’s method is to find the threshold that minimizes the variances of the histogram in each class. This can be shown to be equivalent of maximizing the between class variance, after making some assumptions including the process is stationary. We choose to maximize the between class variance because it can be calculated recursively (Otsu 1979, Sankur 2004, Turkel).
Total variance is the sum of the within class variance and the between class variance. Since the image is stationary, the variance is constant and minimizing one term is equivalent to maximizing the opposing term.
Need to find that maximizes
In class probabilities:
In class means:
In class variances:
Probability mass function:
where, is the maximum luminance value in the image. In my case this value if 255. is number of pixels, and is the number of pixels at value Note:
I implemented this algorithm in two ways. In the first strategy, from a set of 22 images from a single video, I found the Otsu threshold in the first image and then used that threshold for the rest of the images in the set. In the second strategy, I calculated the Otsu threshold for each image and used that threshold.
Adaptive Threshold Methods
In adaptive threshold methods, a unique threshold is found for each pixel in the image using a kernel of a specified block size centered on the pixel of interest. This method effectively smooths the local area before thresholding. OpenCV has two adaptive methods that I ran on my image samples, the mean kernel and the Gaussian kernel (OpenCV 2010).
Mean Kernel
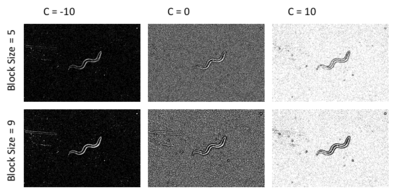
This method computes the local mean in the specified block size. The value of the pixel is above the local mean, the pixel is classified as foreground. If not, it is classified as background. To analyze this method, I ran the algorithm for a variety of block sizes and parameter C (OpenCV 2010).
Gaussian Kernel
In the Gaussian kernel method, the threshold T(x,y) is found by pointwise multiplying a Gaussian window of the specified block size with the neighborhood around the current pixel. The product is them summed over all the points in the neighborhood. If the value of the pixel is above the calculated threshold, it is classified as foreground, otherwise it is classified as background. To analyze this method, I ran the algorithm for a variety of block sizes and parameter C (OpenCV 2010).
Where is a Gaussian window specified by the block size.
chosen such that
Summary of Threshold Methods
The following table gives a summary of the different methods analyzed:
Results:
The Otsu method proved to be the most effective of all threshold methods. The result of the contour finding processing step from three arbitrarily chosen frames are compared in the following figure for methods 1, 2, and 3. In the contour finding step, contours are drawn around all the foreground objects found in the thresholding step and the largest continuous contour is specified at the worm and is indicated by the blue contour. The first method is the empirically chosen threshold of 75 that I started with. In this method, the largest contour was always found to be the noise and cantilever outline in the upper left corner of the image. In the second method, the Otsu threshold was found on the first image (Frame 400) and was used for the rest of the frames in the video. This works under the assumption that the relative histograms of the foreground and background will not change much from frame to frame and time could be saved by not running the calculation on each frame. The Otsu threshold found for the first frame was 115. In the third method, the Otsu threshold was found uniquely for each frame. The average Otsu threshold among the 22 frames was 122.13 with a standard deviation of 12.84.
Methods 2 and 3, which use the Otsu threshold were 100% successful in identifying the largest contour as the worm. However, the contour is not the desired outline of the body of the worm. Instead the side walls of the worm are illuminated creating portions of the worm to be left out and the contour not smooth. In order to use this method, the worm must be more evenly illuminated. This is a key finding from this investigation as it affects the hardware construction of the system. Instead of a single light source from one direction, I will employ a ring light that surrounds the sample, applying light from all directions so that the worm is more evenly illuminated.
The adaptive threshold methods each had two parameters to modulate: the block size and the parameter C. I ran each method with different combinations of both parameters. The results are shown in the figures below. For C = 0, C = 10, the threshold results yielded exactly zero found contours in the thresholded images. Increasing the block size yielded a better outline of the worm, as we will investigate in the next section.
Mean Kernel Results:
Gaussian Kernel Results:
To demonstrate the effectiveness of the adaptive thresholding methods, we must take a look at the results of the contour finding to see the percentage of frames that correctly identified the worm as the largest contour. The Gaussian kernel was much more effective at creating the worm as the largest contour. It makes sense that this method is more accurate because of the way it weights the pixels in the neighborhood. The pixels closest to the center are weighted higher than those farther from the center. It is also remarkable that the score decreases as the block size gets really big. I suspect this is because the filtering becomes too high, effectively smearing the background making other objects such as the cantilever, making that contour bigger that the one surrounding the worm. The smearing also brings out the entire worm body. Therefore, the contour around the worm is a nice outline of the body, as we desire.
While the effectiveness of finding a nice contour of the worm's body is imperative, I must also consider the computation time. In order to process these images in real time, the entire skeleton finding process must be less than 66.6 ms, as governed by the 15 fps rate of the camera. Being well under this processing time is preferred as a buffer. Therefore, the time required to threshold was found for each method. The following figure shows the average time it took to find the thresholded image. The Otsu method requires no significant extra processing time, which makes it a great candidate for this application. Calculating the Otsu threshold hardly increases the threshold finding time! The adaptive threshold techniques are inherently slow because of their two dimensional nature and thus make them poor candidates for this application.
To demonstrate the correlation between computation time and the two dimensional nature of the algorithm, I have plotted the block size versus the computation time for each method in the figure below. The Gaussian kernel method computation time increases linearly with block size, but surprisingly, the mean kernel remains constant. I was surprised by this result, given that the amount of computation to find the threshold is effectively the same in both methods: weight each pixel by some value and then sum the weighted pixels. In the Gaussian kernel method though, we need to calculate what those weights are based on the size of the block, which is increases in complexity as the block size increases. This is the result of the linear increase in threshold finding time with block size that we see in these results.
Finally, given the findings in the threshold investigation, the next version of the hardware is shown below. The key improvement is the use of a ring light surrounding the sample, as opposed to a single light source from one side. This even lighting will better illuminate the entire worm so that the threshold finding algorithm will binarize the entire worm as opposed to just the side walls, creating a nice contour around the body of the worm. Other features of the hardware construction include a structurally sound platform, a movable x, y stage controllable by a computer so that the sample can be moved relative to the cantilever mounted above it. The camera and optics will also be mounted on an automatically controlled jack so that the focus of the camera can be well controlled and maintained. This is sketched in the figure below.
Conclusions:
In this project, I successfully investigated a variety of thresholding techniques, which provided valuable insight into improvements for a second revision of a hardware design for a system to track freely moving C. elegans in real time. The integrated system will apply calibrated forces to the body of the worms with high spatial precision, providing high quality, high throughput data for behavioral analysis. Through this project I learned just how important it is that the front end steps of image processing be done with great care as the success of the downstream steps hinge greatly on the quality of the image and thresholding.
Otsu's method looks at the in class and between class variances of the two sides of a histogram divided by a threshold. The threshold is set such that the between class variance is maximized. In my analysis it effectively separates the worm from the rest of the background. However, the contour of the worm, was not a smooth body outline because the worm's body was only illuminated from one side. As a result, when I implement the next version of the hardware, I will be using a ring light to source light from all directions, as opposed to the original light source design of only a single light source. This is the key conclusion from my findings. Moving forward, I will be using Otsu's Method in my software, paired with a ring light around my sample.
The mean kernel and Gaussian kernel threshold methods both improved the performance of thresholding, but had significant drawbacks. The increased computation time is too high for the available window for the entire skeletonization of the worm. The success rate of finding the worm was also too low. The effective filtering of the image frequently blurred the noise eso much that the contours around it because longer than around the worm itself. The increased block size of the kernels improved preservation of the worm's body outline.
Future work in this project includes first constructing the next, more stable version of the optical system. Once constructed the improved Otsu thresholding method will be implemented, and further investigation into improved methods for tracking the head and tail will be started. Other hardware including an automatic x,y microscope stage and cantilever control will also be integrated for a fully functional system.
References
- Goodman, Miriam B. (2006). "Mechanosensation," "WormBook", http://www.wormbook.org/chapters/www_mechanosensation/mechanosensation.html
- Park, Sung-Jin, Petzold, Bryan C., Goodman, Miriam B., Pruitt, Beth L., (2011). "Piezoresistive cantilever force-clamp system," "Review of Scientific Instruments", 82(2).
- Petzold, Bryan C, Park, Sung-Jin, Mazzochette, Eileen A., Goodman, Miriam B., Pruitt, Beth L. (2013). "MEMS-based Force-clamp Analysis of the Role of Body Stiffness in C. elegans Touch Sensation," "Integrative Biology", In Press.
- O'Hagan, Robert, Chalfie, Martin, Goodman, Miriam B. (2004). "The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals," "Nature Neuroscience", 8(1).
- Leifer, Andrew M, Fang-Yen, Christopher, Gershow, Marc, Alkema, Mark J., Samuel, Aravinthan D. T., (2011). "Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans," "Nature Methods", 8(2).
- Sankur, Bulent, Mehmet, Sezgin. (2004). "Survey over image thresholding techniques and quantitative performance evaluation," "Journal of Electronic Imaging", 13(1).
- Otsu, Nobuyuki (1979). "A Threshold Selection Method from Gray-Level Histograms," IEEE Transactions on Systems, Man and Cybernetics, 9(1).
- Turkel, Eli. "Automatic Thresholding," http://www.math.tau.ac.il/~turkel/notes/otsu.pdf Retrieved 18 March 2013
- Bradski, Gary. "Miscellaneous Image Transformations," "Open CV Wiki," (2010). http://opencv.willowgarage.com/documentation/cpp/miscellaneous_image_transformations.html Retrieved 16 March 2013
Appendix: Code
A note on the code: This code provided here is the implementation function for finding the worm written in C++. It is the highest level function in a complete module of functions. It includes all the threshold functions and contour finding functions provided by the OpenCV library. The other custom functions for finding the head and tail, segmenting and skeletonizing are outside the scope of this project so I did not include them in this appendix. There are also a series of structures used to store the images, data and other parameters that are defined outside of this function, but in my worm analysis module, but have not been included here either. The main function simply loads an image and calls the FindWorm function to find the worm in the image and then writes the data to a csv file and saves the processed image.
void FindWorm(WormImageStructures &WormImages, WormDataStructures &WormData, WormDataStructures &PreviousWormData, WormThresholdParameters &WormThresholdParam, GaussianSmoothingParameters &WormSmoothParam, WormContourFindingParameters &WormContourParam, WormSegmentationStructures &WormSeg, TimingStructures &Timer){
//This Function is the highest level function for finding the skeleton of the worm.
//All the data is passed to the function by reference from the main function.
//Save move worm to previous worm:
if(!WormData.FirstImageFlag){
PreviousWormData = WormData;
}
//Timer initialize:
clock_t start;
//Data for Gaussian smoothing:
Size_<int> ksize(WormSmoothParam.ksizeWidth,WormSmoothParam.ksizeHeight);
//Data for contour finding
vector<Vec4i> hierarchy;
Point offset = (0,0);
//Images for printing:
WormData.ImageToPrint = Mat::zeros(WormImages.OriginalImage.rows, WormImages.OriginalImage.cols, CV_8UC3);
WormData.ImageToPrint2 = Mat::zeros(WormImages.OriginalImage.rows, WormImages.OriginalImage.cols, CV_8UC3);
start = clock();
//THRESHOLDING SECTION
//Method 1:
//threshold( WormImages.OriginalImage, WormImages.ThresholdImage, WormThresholdParam.threshold, WormThresholdParam.maxVal, WormThresholdParam.thresholdType);
//Method 2:
//Threshold to create binary image
//if(WormData.FirstImageFlag){
// WormThresholdParam.OtsuThreshold = threshold( WormImages.OriginalImage, WormImages.ThresholdImage, WormThresholdParam.threshold, WormThresholdParam.maxVal, WormThresholdParam.thresholdType + THRESH_OTSU);
//}
//else {
// threshold( WormImages.OriginalImage, WormImages.ThresholdImage, WormThresholdParam.OtsuThreshold, WormThresholdParam.maxVal, WormThresholdParam.thresholdType);
//}
//Method 3:
//WormThresholdParam.OtsuThreshold = threshold( WormImages.OriginalImage, WormImages.ThresholdImage, WormThresholdParam.threshold, WormThresholdParam.maxVal, WormThresholdParam.thresholdType + THRESH_OTSU);
//Method 4:
//int BlockSize = 55;
//double C = -10;
//adaptiveThreshold(WormImages.OriginalImage, WormImages.ThresholdImage, WormThresholdParam.maxVal, ADAPTIVE_THRESH_MEAN_C, WormThresholdParam.thresholdType, BlockSize, C);
//Method 5:
int BlockSize = 55;
double C = -10;
adaptiveThreshold(WormImages.OriginalImage, WormImages.ThresholdImage, WormThresholdParam.maxVal, ADAPTIVE_THRESH_GAUSSIAN_C, WormThresholdParam.thresholdType, BlockSize, C);
// SMOOTHING AND FINDING CONTOURS
//Smooth using Gaussian Filter
GaussianBlur(WormImages.ThresholdImage, WormImages.SmoothImage, ksize, WormSmoothParam.sigmaX, WormSmoothParam.sigmaY, WormSmoothParam.borderType);
Timer.difThreshold = ( std::clock() - start ) / (double)CLOCKS_PER_SEC;
//Find Contours
findContours(WormImages.SmoothImage, WormData.Contours, hierarchy, WormContourParam.mode, WormContourParam.method, offset);
Timer.difContour = ( std::clock() - start ) / (double)CLOCKS_PER_SEC;
//Find the largest and the second largest contour:
int NumberOfContours = WormData.Contours.size();
int CurrentContourSize;
int MaxContourSize = 1;
int MaxContour=1;
for(int i = 0; i<NumberOfContours; i++){
CurrentContourSize = WormData.Contours[i].size();
if (CurrentContourSize>MaxContourSize){
MaxContour = i;
MaxContourSize = CurrentContourSize;
}
}
WormData.Worm = WormData.Contours[MaxContour];
//FINDING HEAD AND TAIL
FindWormTail(WormData);
Timer.difTail = ( std::clock() - start ) / (double)CLOCKS_PER_SEC;
FindWormHead(WormData);
Timer.difHead = ( std::clock() - start ) / (double)CLOCKS_PER_SEC;
//If this is not the first image, check against previous head and tail.
if(!WormData.FirstImageFlag){
HeadTailCheck(WormData, PreviousWormData);
}
//SEGMENTATION
// Perform the segmentation of the worm:
WormSegmentation(WormData, WormSeg);
Timer.difSegmentation = ( std::clock() - start ) / (double)CLOCKS_PER_SEC;
//FIND SKELETON
//Use the segments to find the skeleton of the worm:
FindSkeleton(WormData, WormSeg);
Timer.difSkeleton = ( std::clock() - start ) / (double)CLOCKS_PER_SEC;
//Now done with at least one image, so turn off first image flag.
WormData.FirstImageFlag = false;
}










![{\displaystyle \sigma _{b}^{2}(T)=P_{f}(T)[1-P_{f}(T)][\mu _{f}(T)-\mu _{b}(T)]^{2}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/13c76db4e10a646d93cbbb5766678dc9e133b9e0)




![{\displaystyle \sigma _{f}^{2}=\sum _{g=0}^{T}{[g-\mu _{f}(T)]^{2}{\frac {p(g)}{P_{f}(T)}}},\sigma _{b}^{2}=\sum _{g=T+1}^{G}{[g-\mu _{b}(T)]^{2}{\frac {p(g)}{P_{b}(T)}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/91609940fbf4ec6827b8abac8911df982e9956c0)